6.5 EMP_dimension_analysis
The module EMP_dimension_analysis supports multidimensional data dimensionality reduction analysis, with dimensionality reduction methods including Principal Component Analysis (PCA), Principal Coordinates Analysis (PCoA), Partial Least Squares (PLS), and Orthogonal Partial Least Squares (OPLS).
6.5.1 Analysis and visualization of microbial beta diversity
If PCA dimensionality reduction is required, simply set the
method parameter to 'pca'.
🏷️Example:
Extract the assay of taxonomy from the MAE object. Utilize the module EMP_identify_assay to screen the core microbial data. Use the module EMP_collapse to collapse the core microbial data into genus-level. And perform a PCoA dimensionality reduction analysis with the module EMP_dimension_analysis.
MAE |>
EMP_assay_extract('taxonomy') |>
EMP_identify_assay(method='default',estimate_group = 'Group') |>
EMP_collapse(estimate_group = 'Genus',collapse_by = 'row') |>
EMP_dimension_analysis(method = 'pcoa',distance='bray')
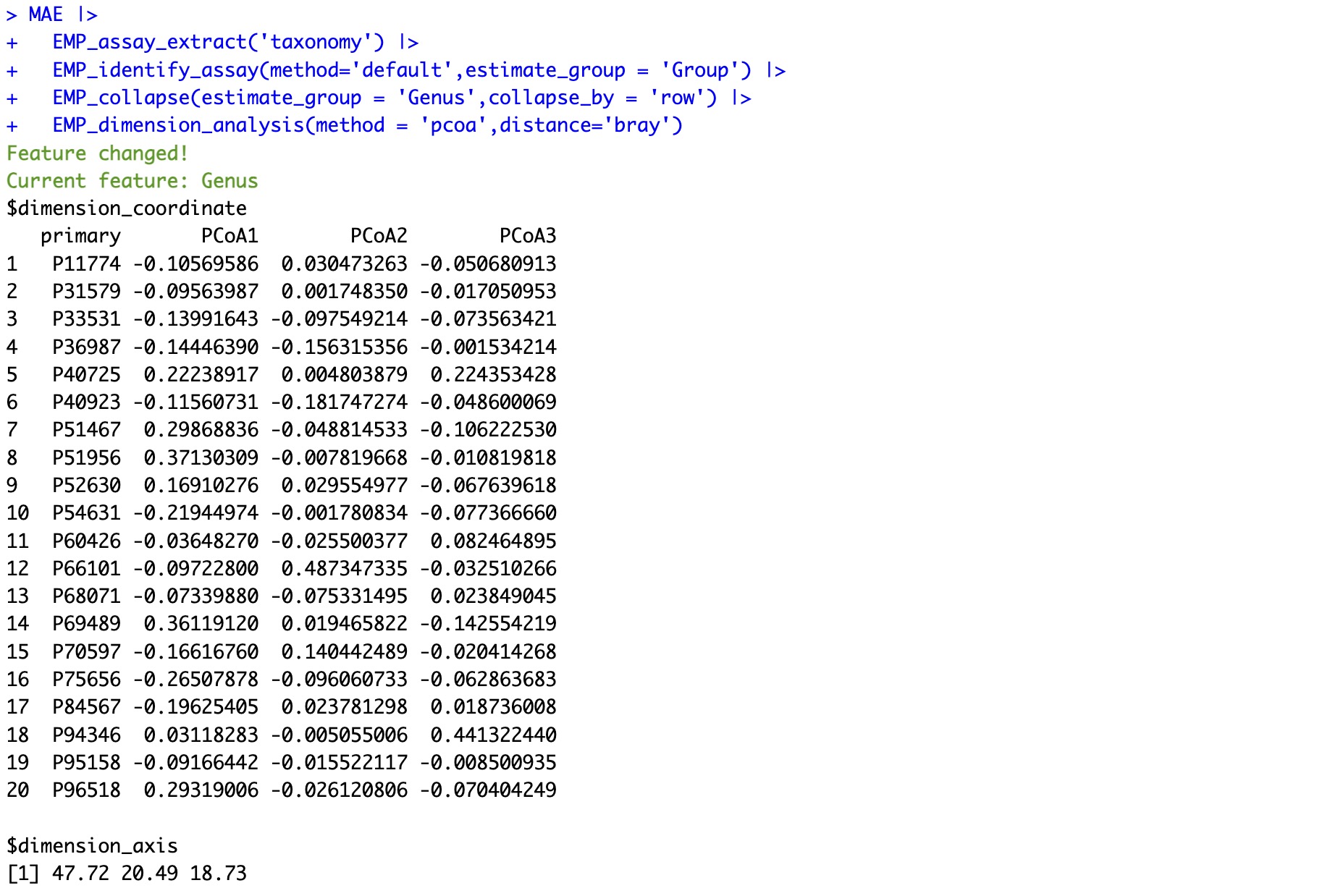
The results of dimensionality reduction can be visualized using module EMP_scatterplot .
The parameter
show supports the presentation of dimensionality reduction results from three dimensions: p12, p13, and p23, and provides an interactive HTML graphic.
MAE |>
EMP_assay_extract('taxonomy') |>
EMP_identify_assay(method='default',estimate_group = 'Group') |>
EMP_collapse(estimate_group = 'Genus',collapse_by = 'row') |>
EMP_dimension_analysis(method = 'pcoa',distance='bray') |>
EMP_scatterplot(estimate_group='Group',show='p12html',ellipse=0.3)
6.5.2 Metabolomics data PLS/OPLS dimensionality reduction analysis and feature metabolites screening
If OPLS is required, simply set the
method parameter to 'opls'.
🏷️Example:
Extract the assay of untarget_metabol from the MAE object, collapse the data according to secondary metabolites annotated with KEGG, perform relative abundance transformation, conduct PLS dimensionality reduction analysis, obtain the reduced dimension coordinates and VIP values of the metabolites, and filter the characteristic metabolites based on the VIP values.
MAE |>
EMP_assay_extract(experiment = 'untarget_metabol') |>
EMP_collapse(estimate_group = 'MS2kegg',collapse_by='row',
na_string = c("NA", "null", "","-"),
method = 'mean',collapse_sep = '+') |>
EMP_decostand(method = 'relative') |>
EMP_dimension_analysis(method = 'pls',estimate_group = 'Group') |>
EMP_filter(feature_condition = VIP >2)

6.5.3 UMAP (Uniform Manifold Approximation and Projection)
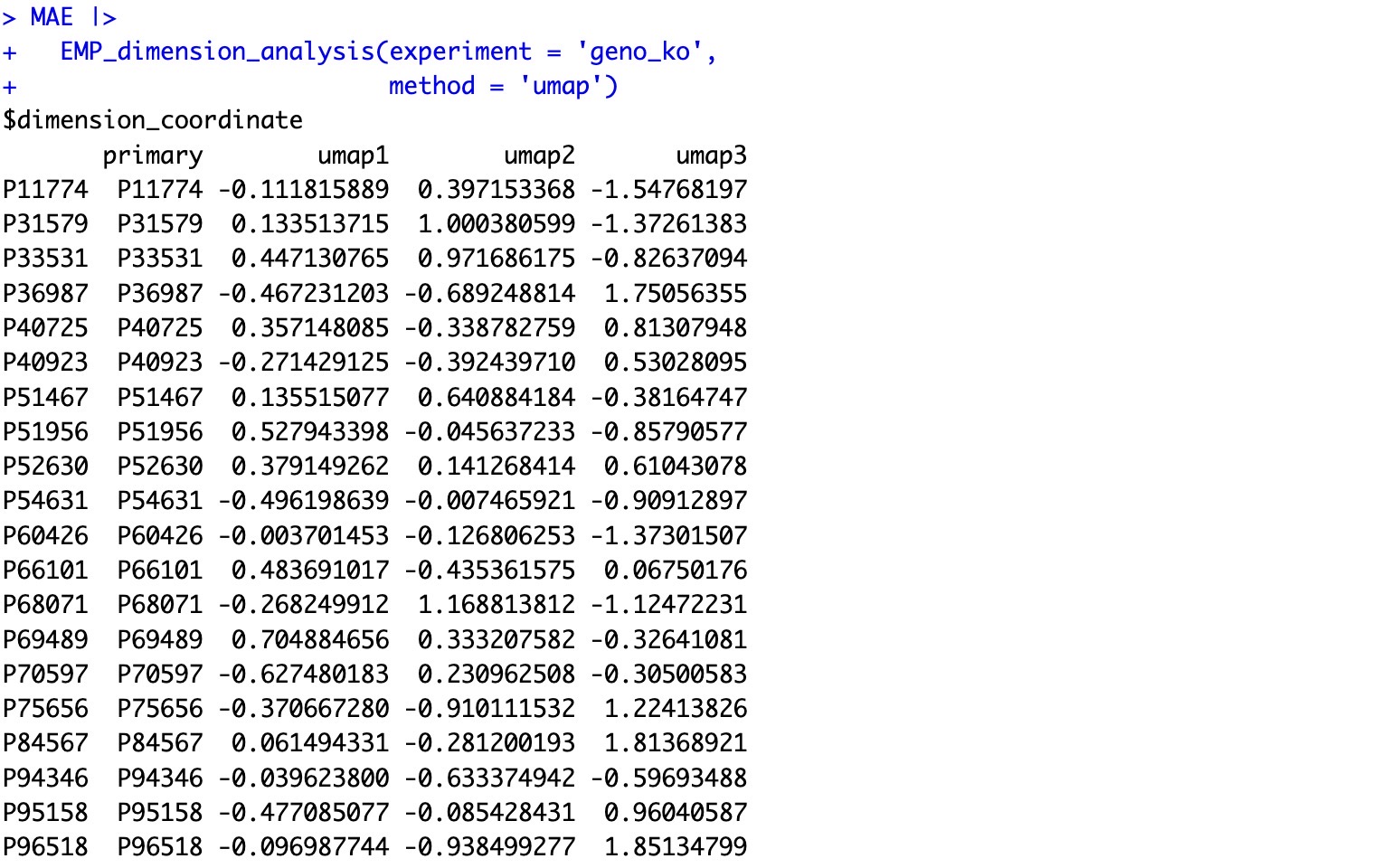
🏷️Example1: Make the UMAP analysis based on KO data
MAE |>
EMP_dimension_analysis(experiment = 'geno_ko',
method = 'umap')
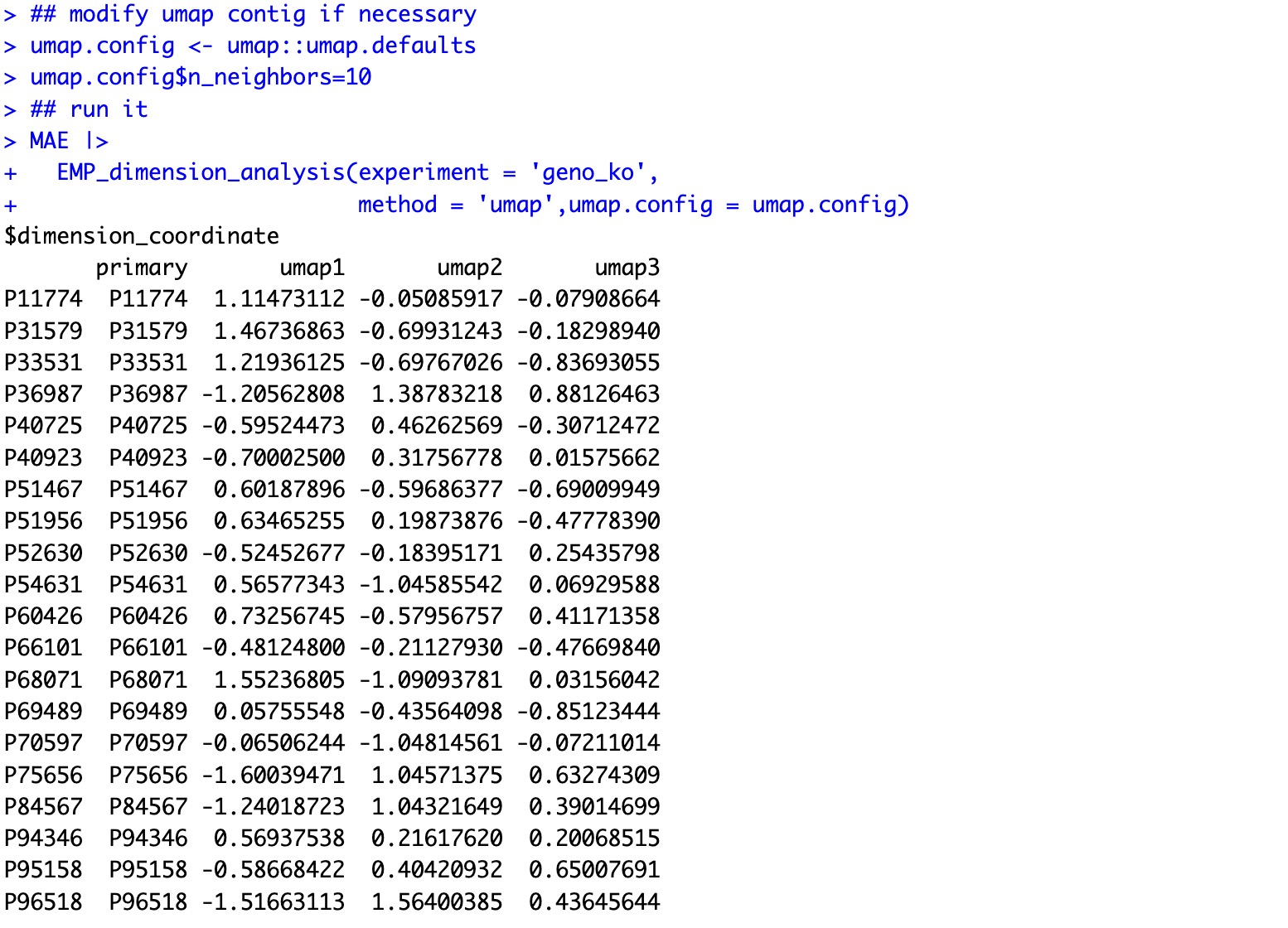
🏷️Example2: Modify the parameter for UMAP
Get the parameter file from the umap package and change it.
umap.config <- umap::umap.defaults
umap.config$n_neighbors <- 10
Input the parameter in the module.
MAE |>
EMP_dimension_analysis(experiment = 'geno_ko',
method = 'umap', umap.config = umap.config)